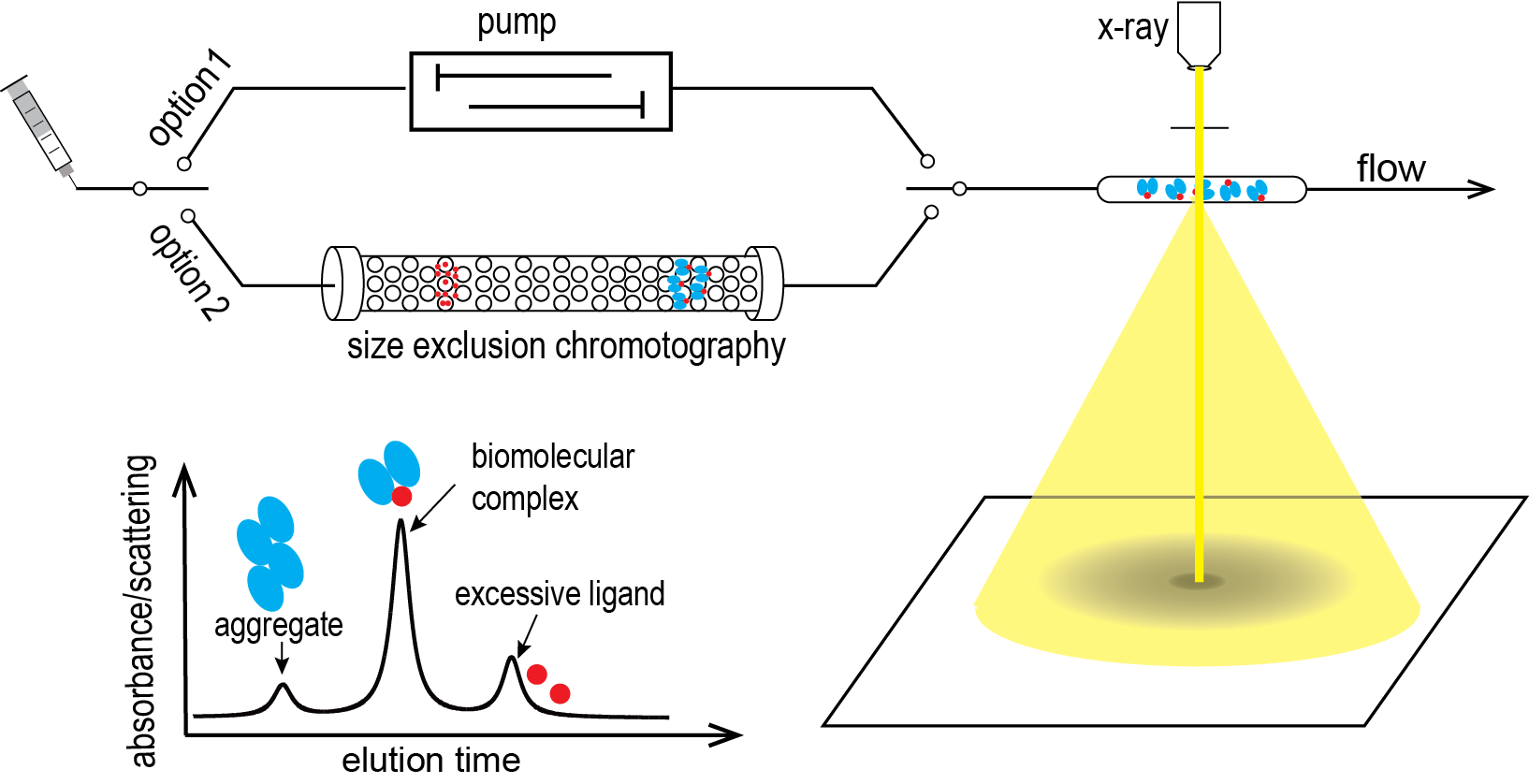
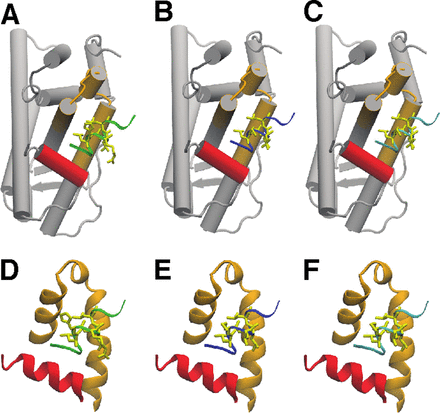
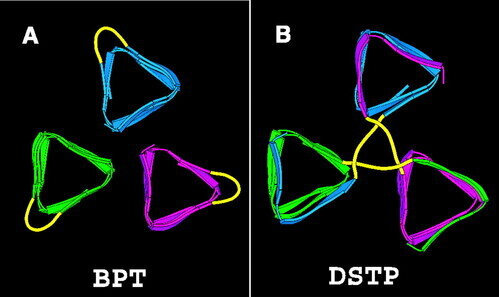
Selected Publications
Targeting the ERα DBD-LBD interface with mitoxantrone disrupts receptor function through proteasomal degradation
Han Wang, Yuxuan Luo, Sandeep Artham, Qianqian Wang, Yi Peng, Zixi Yun, Xinyue Li, Chen Wu, Zhenghao Liu, Kristen L. Weber-Bonk, Chun-Peng Pai, Yuan Cao, Jiangan Yue, Sunghee Park, Ruth A. Keri, Lisheng Geng, Donald P. McDonnell, Hung-Ying Kao, and Sichun Yang
Molecular Cancer Therapeutics, in press (2025) (DOI: 10.1158/1535-7163.mct-25-0405)
Abstract: The estrogen receptor (ER or ERα) remains the primary therapeutic target for luminal breast cancer, with current treatments centered on competitive antagonists, receptor down-regulators, and aromatase inhibitors. Despite these options, resistance frequently emerges, highlighting the need for alternative targeting strategies. We discovered a novel mechanism of ER inhibition that targets the previously unexplored interface between the DNA-binding domain (DBD) and ligand-binding domain (LBD) of the receptor. Through computational screening and functional assays, we identified mitoxantrone (MTO), an FDA-approved topoisomerase II inhibitor, as a specific ligand for this DBD-LBD interface. Comprehensive biophysical, biochemical, and cellular analyses demonstrate that MTO binding induces distinct conformational changes in ER, triggering rapid cytoplasmic redistribution and proteasomal degradation through mechanisms independent of its DNA damage activity. Critically, MTO effectively inhibits constitutively active ER mutants (Y537S and D538G) associated with endocrine therapy resistance, suppressing both wild-type and mutant ER-dependent gene expression and tumor growth more potently than fulvestrant in cellular and xenograft models. These findings establish the DBD-LBD interface as a druggable allosteric site that can overcome conventional resistance mechanisms, providing a new therapeutic paradigm for targeting nuclear receptor function through disruption of interdomain communication rather than hormone binding competition.
The sequence-structure-function relationship of intrinsic ERα disorder
Zhanwen Du, Han Wang, Shuqi Luo, Zixi Yun, Chen Wu, Wangfei Yang, Matthias Buck, Wenwei Zheng, Alexandar L. Hansen, Hung-Ying Kao, and Sichun Yang
Nature, 638, 1130–1138 (2025) [https://www.nature.com/articles/s41586-024-08400-1]
Hydrophobic driving of disordered proteins
The estrogen receptor (ER or ERα), a nuclear hormone receptor that drives most breast cancer, is commonly activated by phosphorylation at serine 118 within its intrinsically disordered N-terminal transactivation domain. Although this modification enables estrogen-independent ER function, its mechanism has remained unclear despite ongoing clinical trials of kinase inhibitors targeting this region. By integration of small-angle X-ray scattering and nuclear magnetic resonance spectroscopy with functional studies, we show that serine 118 phosphorylation triggers an unexpected expansion of the disordered domain and disrupts specific hydrophobic clustering between two aromatic-rich regions. Mutations mimicking this disruption rescue ER transcriptional activity, target-gene expression and cell growth impaired by a phosphorylation-deficient S118A mutation. These findings, driven by hydrophobic interactions, extend beyond electrostatic models and provide mechanistic insights into intrinsically disordered proteins, with implications for other nuclear receptors. This fundamental sequence–structure–function relationship advances our understanding of intrinsic ER disorder, crucial for developing targeted breast cancer therapeutics.
News and Media Coverage:
Searching for Protein Off-Targets of Prostate-Specific Membrane Antigen-Targeting Radioligands in the Salivary Glands
William Julian, Olga Sergeeva, Wei Cao, Chunying Wu, Bernadette Erokwu, Chris Flask, Lifang Zhang, Xinning Wang, James Basilion, Sichun Yang, Zhenghong Lee
Cancer Biotherapy & Radiopharmaceuticals, 39 (10), 721-732 (2024)
Phosphorylation modulates estrogen receptor disorder by altering hydrophobic clustering of residual structures
Zhanwen Du, Han Wang, Chen Wu, Matthias Buck, Wenwei Zheng, Alexandar L. Hansen, Hung-Ying Kao, Sichun Yang
BioRxiv preprint (2023) —- for the record
Protein intrinsic disorder is coupled to a range of biological phenomena, from gene regulation to cancer progression. A pivotal example is the phosphorylation of the estrogen receptor (ER) at Ser118 in its N-terminal domain (NTD), resulting in the activation of its transcriptional function. Owing to the complex nature of its intrinsic disorder, however, the precise mechanism underpinning how this modification regulates ER activity remains elusive. Conventionally, the mechanistic effect of Ser118 phosphorylation was thought to be driven primarily by electrostatic interactions by the negative charges originating from the phosphate group. Instead, using biophysical approaches of small-angle X-ray scattering and nuclear magnetic resonance spectroscopy, we herein demonstrate that Ser118 phosphorylation triggers long-range conformational changes in the disordered ER-NTD, altering the hydrophobic clustering of residual structures and favoring a more open conformational ensemble of its activated state. Alanine substitutions of hydrophobic amino acids near Ser118 produce similar conformational alterations and rescue the impaired ER activity caused by a phosphorylation-deficient mutant. Contrary to the conventional notion, these findings lead to a phenomenon that the modulation of hydrophobic clustering is a key factor in activating the transcriptional function, regardless of whether there is electrostatic repulsion arising from Ser118 phosphorylation. These findings establish a new structure-function paradigm for this disordered protein by directly connecting phosphorylation-induced conformational changes with its transcriptional functionality.
Biophysical and Integrative Characterization of Protein Intrinsic Disorder as a Prime Target for Drug Discovery
Shuqi Luo, Samuel Wohl, Wenwei Zheng, and Sichun Yang
Biomolecules 2023, 13(3), 530 (brief review)
[DOI]
Protein intrinsic disorder is increasingly recognized for its biological and disease-driven functions. However, it represents significant challenges for biophysical studies due to its high conformational flexibility. In addressing these challenges, we highlight the complementary and distinct capabilities of a range of experimental and computational methods and further describe integrative strategies available for combining these techniques. Integrative biophysics methods provide valuable insights into the sequence–structure–function relationship of disordered proteins, setting the stage for protein intrinsic disorder to become a promising target for drug discovery. Finally, we briefly summarize recent advances in the development of new small molecule inhibitors targeting the disordered N-terminal domains of three vital transcription factors.
Incorporation of D2O-induced Fluorine Chemical Shift Perturbations into Ensemble-Structure Characterization of the ERalpha Disordered Region
Wenwei Zheng, Zhanwen Du, Soo Bin Ko, Nalinda Wickramasinghe, and Sichun Yang
J. Phys. Chem. B, 2022 126 (45): 9176-9186
[DOI]
Structural characterization of intrinsically disordered proteins (IDPs) requires a concerted effort between experiments and computations by accounting for their conformational heterogeneity. Given the diversity of experimental tools providing local and global structural information, constructing an experimental restraint-satisfying structural ensemble remains challenging. Here, we use the disordered N-terminal domain (NTD) of the estrogen receptor alpha (ERalpha) as a model system to combine existing small-angle X-ray scattering (SAXS) and hydroxyl radical protein footprinting (HRPF) data and newly acquired solvent accessibility data via D2O-induced fluorine chemical shifting (DFCS) measurements. A new set of DFCS data for the solvent exposure of a set of 12 amino acid positions were added to complement previously acquired HRPF measurements for the solvent exposure of the other 16 nonoverlapping amino acids, thereby improving the NTD ensemble characterization considerably. We also found that while choosing an initial ensemble of structures generated from a different atomic-level force field or sampling/modeling method can lead to distinct contact maps even when the same sets of experimental measurements were used for ensemble-fitting, comparative analyses from these initial ensembles reveal commonly recurring structural features in their ensemble-averaged contact map. Specifically, nonlocal or long-range transient interactions were found consistently between the N-terminal segments and the central region, sufficient to mediate the conformational ensemble and regulate how the NTD interacts with its coactivator proteins.
Fusion‐Associated Carcinomas of the Breast: Diagnostic, Prognostic, and Therapeutic Significance
Suet Kee Loo, Megan E. Yates, Sichun Yang, Steffi Oesterreich, Adrian V. Lee, Xiao-Song Wang
Genes, Chromosomes and Cancer, 2022 61:261–273
[DOI]
Integrative structural modeling of a multidomain polo-like kinase
Hao Ruan, Janna Kiselar, Weilin Zhang, Siyang Li, Ruoyao Xiong, Ying Liu, Sichun Yang and Luhua Lai
Phys. Chem. Chem. Phys., 2020, 22, 27581-27589
[DOI]
[Editorial Overview]
Integrative Biophysics: Protein Interaction and Disorder
Sichun Yang and Pau Bernadó
Journal of Molecular Biology 432 (2020) pp. 2843-2845
Over the last decades, individual biophysical techniques and their complementary data integrations have been pivotal to advance functional and disease-driven understanding of biomolecules and their dynamics …
Glycine substitution in SH3-SH2 connector of Hck tyrosine kinase causes population shift from assembled to disassembled state
Lei Huang, Michelle Wright, Sichun Yang, Lydia Blachowicz, Lee Makowski, Benoît Roux
Biochimica et Biophysica Acta (BBA) 1864, 129604 (2020)
[DOI]
Protein Footprinting: Auxiliary Engine to Power the Structural Biology Revolution
Mark R. Chance, Erik R. Farquhar, Sichun Yang, David T. Lodowski, Janna Kiselar
Journal of Molecular Biology 432 (9) 2973-2984 (2020)
[DOI]
A metastable contact and structural disorder in the estrogen receptor transactivation domain
Yi Peng, Shufen Cao, Jana Kiselar, Xiangzhu Xiao, Zhanwen Du, An Hsieh, Soobin Ko, Yinghua Chen, Prashansa Agrawal, Wenwei Zheng, Wuxian Shi, Wei Jiang, Lin Yang, Mark Chance, Witold Surewicz, Matthias Buck, and S. Yang
Structure 27 (2) 229-240 (2019)
[PDF] [HTML]
Featured in a commentary preview by Manalastas and Svergun: Molecular Dissection of the Intrinsically Disordered Estrogen Receptor Alpha-NTD, Structure 27 (2) 207-208 (2019)
Reactive Cysteine Residues in the Oxidative Dimerization and Cu2+ Induced Aggregation of Human D-Crystallin: Implications for Age-Related Cataract
Srinivasagan Ramkumar, Xingjun Fan, Benlian Wang, Sichun Yang, and Vincent Monnier
BBA - Molecular Basis of Disease 1864 3595-3604, (2018)
Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains
Wei Huang, Yi Peng, Janna Kiselar, Xuan Zhao, Aljawharah Albaqami, Daniel Mendez, Ying-Hua Chen, Srinivas Chakravarthy, Sayan Gupta, Corie Ralston , Hung-Ying Kao, Mark Chance, and S. Yang
Nature Communications 9 3520, (2018)
[PDF] [HTML] [Supplementary]
Press release and coverage: EurekAlert, Argonne National Laboratory, Case Daily
Cyclin-dependent kinase 7 (CDK7)-mediated phosphorylation of the CDK9 activation loop promotes P-TEFb assembly with Tat and proviral HIV reactivation
Uri Mbonye, Benliang Wang, Giri Gokulrangan, Wuxian Shi, Sichun Yang, and Jonathan Karn
J. Biol. Chem. 293 10009-10025, (2018)
Charge Neutralization Drives the Shape Reconfiguration of DNA Nanotubes
Pi Liu, Yan Zhao, Xiaoguo Liu, Jixue Sun, Dede Xu, Yang Li, Qian Li, Lihua Wang, Sichun Yang, Chunhai Fan, Jianping Lin
Angewandte Chemie 57 5418-5422, (2018)
A practical guide to iSPOT modeling: An integrative structural biology platform
Advances in Experimental Medicine and Biology Vol. 1009, Chapter 14 (2017),
An Hsieh, Lanyuan Lu, Mark Chance, and S. Yang
SAXS-driven Computational Modeling of Biomolecular Complexes
Synchrotron Radiation in Materials Science, Wiley-VCH Verlag, Chapter 17 (2017)
L. Song, L. Lu, W. Huang, K.M. Ravikuma, J. Meng, S. Yang
[PDF] [HTML]
A Newfound Cancer-activating Mutation Reshapes the Energy Landscape of Estrogen-binding Domain
Journal of Chemical Theory and Computation, 10 (8) 2897–2900, (2014)
Wei Huang, Krishna M Ravikumar, and S. Yang
A simple energy model of coarse-graining predicts the large-scale switching between two well-defined conformations.
[PDF] [HTML] [Supporting Info] [Supporting Movie]
Identification of a novel LXXLL motif in alpha actinin 4 (ACTN4) spliced isoform that is critical for its interaction with estrogen receptor alpha and co-activators
J. Biol. Chem. 287 (42), 35418-35429, (2012)
Simran Khurana, Sharmistha Chakraborty, Xuan Zhao, Yu Liu, Dongyin Guan, Minh Lam, Wei Huang, S. Yang and Hung-Ying Kao
[PDF] [HTML]
Multidomain Assembled States of Hck Tyrosine Kinase in Solution
Proc. Natl. Acad. Sci. USA 107, 15757-15762 (2010)
Sichun Yang, Lydia Blachowicz, Lee Makowski, and Benoît Roux
[PDF] [HTML]
Featured in a News & Views article by Bernadó and Blackledge in Nature (2010).
Structural biology: Proteins in dynamic equilibrium
Nature 468, 1046-1048 (2010)
[PDF] [HTML]
RNA Structure Determination Using SAXS Data
J. Phys. Chem. B. 114 (31), 10039-10048 (2010)
Sichun Yang, Marc Parisien, François Major, and Benoît Roux
[PDF] [HTML]
Note: A webserver of SAXS computing is available at fast-SAXS-pro
A rapid coarse residue-based computational method for X-ray solution scattering characterization of protein folds and multiple conformational states of large protein complexes
Biophysical Journal 96, 4449-4463 (2009)
Sichun Yang, Sanghyun Park, Lee Makowski, and Benoît Roux
[PDF] [HTML] [DOI]
Note: A webserver of SAXS computing is available at fast-SAXS-pro, with new features added to Fast-SAXS, developed for complexes formed by protein and RNA/DNA
Mapping the conformational transition in Src activation by cumulating the information from multiple molecular dynamics trajectories
Proc. Natl. Acad. Sci. USA 106, 3776-3781 (2009)
Sichun Yang, Nilesh K. Banavali, and Benoît Roux
[HTML] [PDF] [Animation]